Spatial omics is making waves across biomedical research, offering transcript-level resolution while preserving spatial context. Whether you’re working in oncology, neuroscience, or developmental biology, spatial omics promises powerful insights—but only if it’s a match for your scientific and operational goals. So, let’s break it down.
❓ Key Questions to Ask Before Starting the Project
- What biological questions are you trying to answer? Is spatial context essential?
- Are you looking at a single tissue or doing comparative multi-tissue studies?
- Do you need whole-transcriptome profiling or targeted panels?
- Will you be working with fresh-frozen, FFPE, or specialized samples?
- Are your collaborators equipped for spatial data analysis?
Asking the right questions can determine if you need ultra-high resolution imaging, single-cell specificity, or just neighborhood-level context. Additional information comparing different techniques can be found in an earlier article Spatial omics overview.
🧪 How to select the appropriate samples?
Not all samples are created equal—spatial omics platforms have specific requirements:
- Fresh-Frozen Tissue: Compatible with most spatial platforms including 10x Genomics Visium and Bruker’s.
- FFPE Samples: Widely used in clinical settings; newer technologies like Xenium (10x) support them.
Prepare your samples early—poor preservation or incompatible prep can waste thousands.
💰 Budgeting for spatial omics
Spatial omics isn’t cheap, and budgeting goes beyond the cost of reagents:
- Equipment & reagent: Special equipment and trained professionals are needed to perform sample processing and sample analysis.
- CRO: Organization that lack in-house resources will need CROs to conduct such a study.
- Data management and analysis: Spatial omics produces various files for sequencing, proteomics and imaging. In order to work with large datasets, bioinformaticians with experience in the cloud services and ability to analyze spatial and pathology data are required.
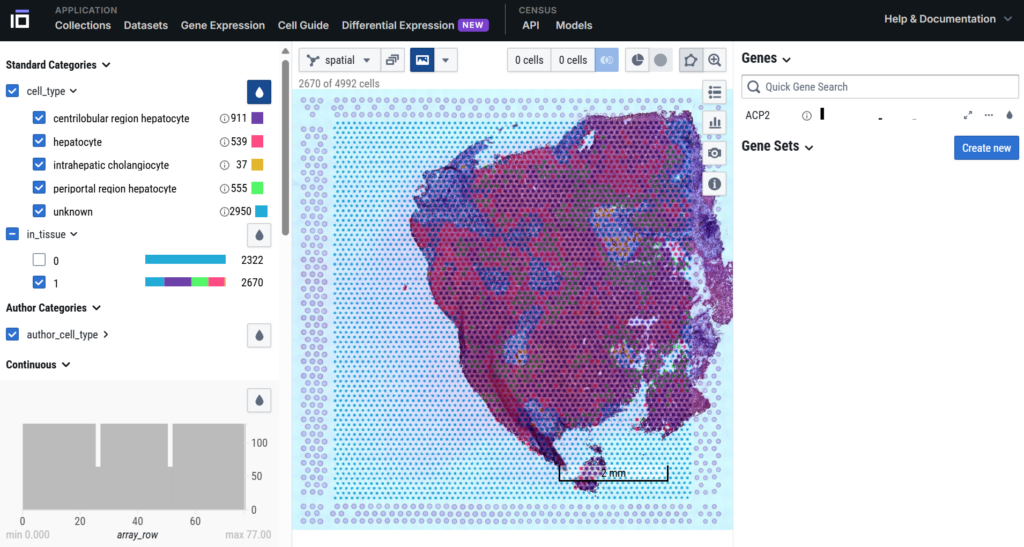
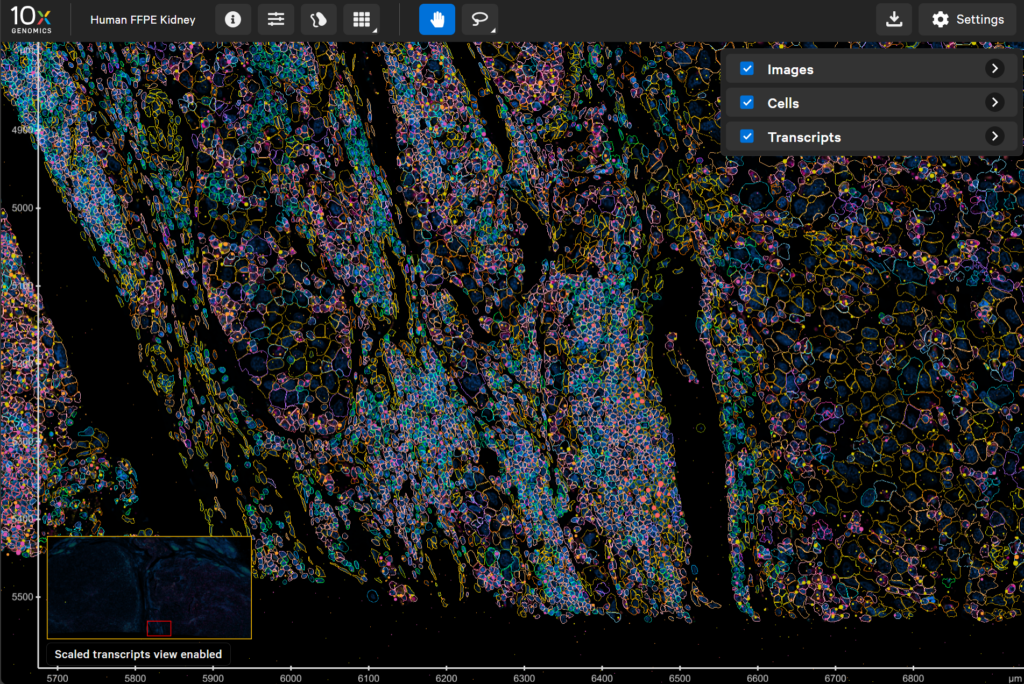
📁 Example datasets to explore
Public datasets can be helpful to evaluate example output data, test pipelines or spark inspiration. Here are a few from 10x genomics and ChanZukerberg Initiative.
- 10x datasets: This provides both Visium and Xenium dataset with option for data exploration within the browser. Datasets – 10x Genomics
- ChanZukerberg Initiative: It provide cross vendor datasets for Visium and MERFISH, with regular additions. Datasets – CZ CELLxGENE Discover
Even just playing with one of these can clarify whether your team is ready for the spatial omics leap.

Leave a Reply