Recently I came across a blog by Cell Signaling Technology describing the InTraSeq assay. What caught my attend was its innovative approach to characterize transcripts and proteins abundance at the single cell resolution. This approach combines single cell RNAseq (scRNAseq) along with antibody-based protein quantitation.
Unlike prior approaches which only detects surface proteins, InTraSeq provides a comprehensive picture for studying both intracellular and extracellular proteins. Thus, one can investigate both the expression of a cell surface receptor as well as modulation of intracellular signaling pathways. The figure below compares the protein abundance with mRNA abundance for a panel of proteins.
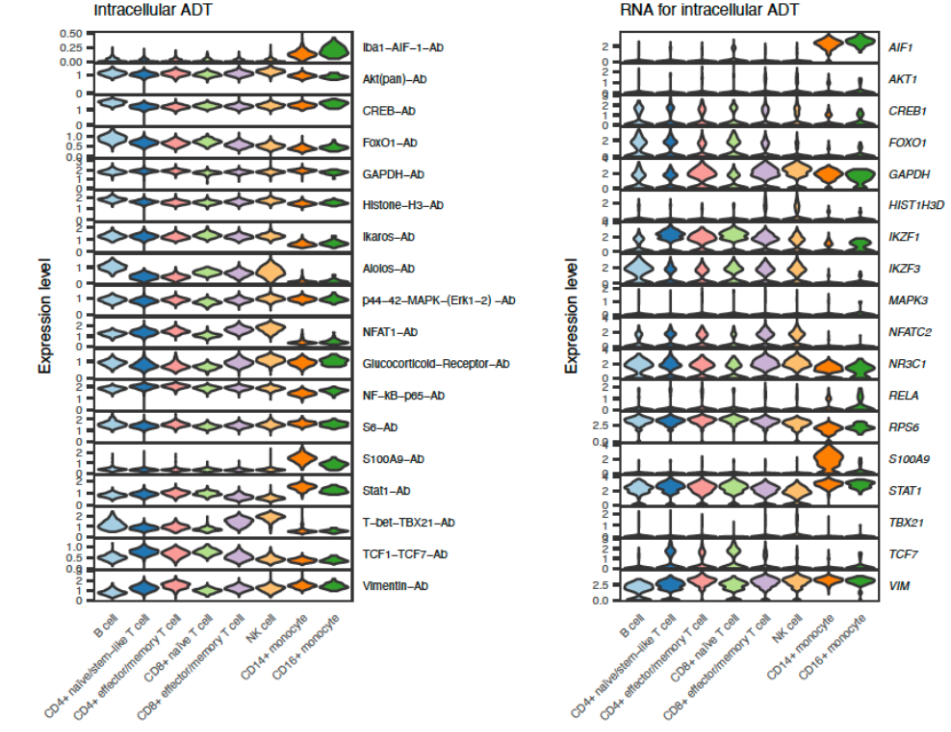
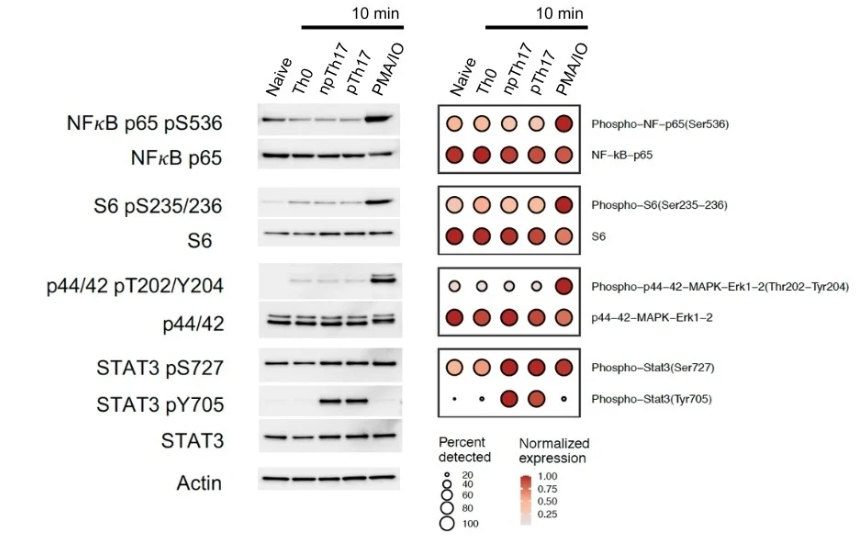
While comparing the results between traditional approaches and InTraSeq, the protein quantitation from the new assay at the single cell level was comparable to those from traditional western blots.
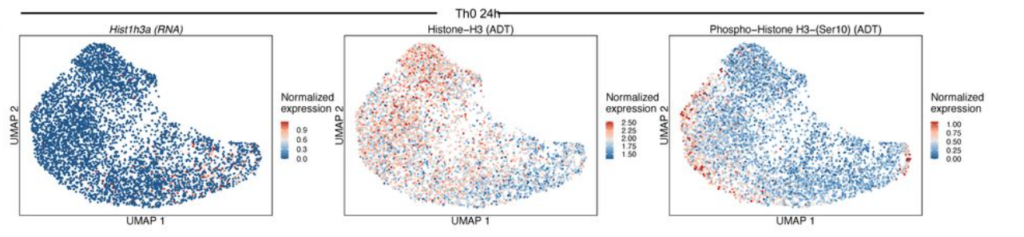
In addition to quantifying whole proteins, it’s possible to detect changes in post translational status. In the figure below the transcript expression of Histone H3 was compared to the total protein abundance and its phospho-Ser10 modification.
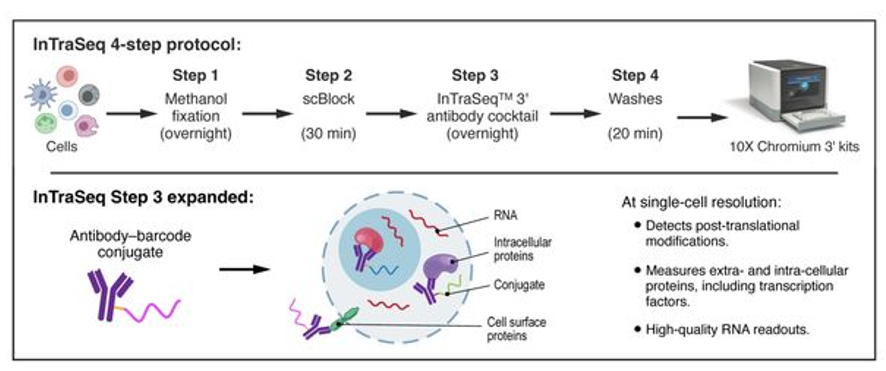
How does it work ?
To detect and quantify proteins the cells are fixed with methanol and then conjugated with antibody-oligo cocktail. Following that the cells are processed by the regular 10X chromium protocol for scRNAseq. Thus, the same study can provide both mRNA and protein abundance for the same cells.
Use cases
- Protein detection and quantitation at both intracellular and cell surface level.
- Evaluate signaling changes at single cell resolution upon stimulation using various agents. Instead of measuring protein in the bulk samples using western or Eliza assay, now it is possible to get granular information for different cell types.
- Ideal for screening drugs that impact proteins and/or their post-translational modification.
Limitations
- Needs high quality validated antibodies for protein quantitation
- Optimization of the technique may be needed for different sample and cell type
Acknowledgement
The figures for this post were obtained from the blog published by Cell Signaling Technology.
References

Leave a Reply